Dask Tips and Tricks – Parallelize a For Loop
Oct 21, 2019
If you're in the scientific computing space there's a good chance you've used Python. A relatively recent addition to the family of awesome libraries in Python for Scientific Computing is Dask. It is a super cool library that allows you to parallelize your code with a very simple and straightforward syntax. There a few aspects of this library that especially call to me.
In no particular order, here they are!
- It can be dropped into an existing codebase with little to no drama! Dask is meant to wrap around existing code and simply decide what can be executed asyncronously.
- Dask can scale either on your laptop or to an entire compute cluster. Without writing MPI code! How cool is that?
- Dask can parallelize data structures we already know and love, such as numpy arrays and data frames.
For those of you sitting here saying, but Spark can do all that, why yes, it can, but I don't find Spark nearly as easy to drop into an existing codebase as Dask. Also, I have I like to learn new things disease!
Let's Execute some Async Code
The least cool, but probably most widely use case I come across for speeding up code is simply parallelizing a for loop. Sure, it's kind of low hanging fruit, but it's easy and you can impress everyone with your code optimization prowess!
For the record, I completely got this from the Dask Tutorial on Github, but since when I googled 'Parallelize a for loop with Dask' nothing quite idiot-proof enough for me came up here we are!
If you want to follow along on your own, scroll down to the bottom to get the source code along with a preconfigured docker instance.
Import Libraries
First of all, let's import the libraries we will be using.
The dask is imported for obvious reasons, while time and timeit are imported for demonstration purposes that will be made clear below.
Our For Loop
In the real world, normally I have some code, that hopefully works, and I get the job of speeding it up. Let's take an example from the Dask Tutorial.
What we have here is a very straightforward for loop. Loop through some data, do some stuff, get some results, and prosper.
How to think about parallelizing
A VERY important point to make here, and one that is important for anyone wanting to get into parallel computing, is to think of your code as a series of tasks. Once you start to think that way you can think about how your tasks are executed, which tasks are dependent on others, and which can be run indepedently.
I may be a computer person, but I always have a stack of graph paper and some nice sparkly pens sitting next to me all the time. If you're unsure of your tasks take the time to draw them out! It really helps! When you draw out your dependency and execution order you should get something that looks like this. In this example task-1 must complete before task-2 can begin.
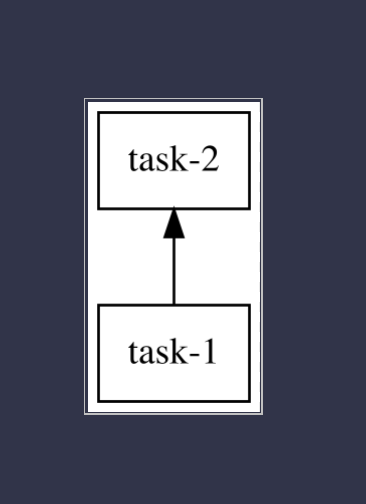
In this particular case, each iteration of the for loop does a computation that can be done completely in isolation of the others. We are appending some items to the results array, but since we are just summing them up at the end the order of the results array doesn't matter.
How to Parallelize with Dask
How that we know which portion of the code can be executed in parallel, we simply tell Dask which tasks to execute asynchronously. Dask uses a keyword called delayed, meaning delay the execution and let the Dask scheduler handle it. Cool, right?!?
For those of you that like decorators, the dask.delayed is also available as a decorator. Check out the docs page on the decorator here.
Test it out!
Now that we have our sequential execution and our async execution mapped out let's try them out and see!
Execute the Sequential Code
If you look closely at the sequential code you will see that we are running 8 iterations at 1 second each due to the sleep function. The whole purpose of the sleep function here is to give us something to compare to.
Execute the Async Code
Take note here that when we execute the async code we have an extra step, compute, where dask actually runs the code. This is because we told dask we have delayed code, meaning wait, so then we must wait until dask gets the full picture so it can decide the most efficient way to execute our tasks.
Check out the Results!
We can see here, that as expected, the sequential code took just about 8 seconds. The dask code took around 2 seconds, and if I was willing to devote more CPUs to my docker instances it could have been even faster.
Get some Code!
As always, all the source code for this example, plus a fully functional docker instance, is available for your to use, completely for free, from my Resource Library.

