Manage High Content Screening CellProfiler Pipelines with Apache Airflow
May 31, 2020
If you are running a High Content Screening Pipeline you probably have a lot of moving pieces. As a non exhaustive list you need to:
- Trigger CellProfiler Analyses, either from a LIMS system, by watching a filesystem, or some other process.
- Keep track of dependencies of CellProfiler Analyses - first run an illumination correction and then your analysis.
- If you have a large dataset and you want to get it analyzed sometime this century you need to split your analysis, run, and then gather the results.
- Once you have results you need to decide on a method of organization. You need to put your data in a database and set up in depth analysis pipelines.
These tasks are much easier to accomplish when you have a system or framework that is built for scientific workflows.
If you prefer to watch I have a video where I go through all the steps in this tutorial.
Enter Apache Airflow
Apache Airflow is :
Airflow is a platform created by the community to programmatically author, schedule and monitor workflows.
There are a ton of great introductory resources out there on Apache Airflow, but I will very briefly go over it here.
Apache Airflow gives you a framework to organize your analyses into DAGs, or Directed Acyclic Graphs. If you aren't familiar with this term it's really just a way of saying Step3 depends upon Step2 which depends upon Step1, or Step1 -> Step2 -> Step3.
Apache Airflow uses DAGs, which are the bucket you throw you analysis in. Your DAG is comprised of Operators and Sensors. Operators are an abstraction on the kind of task you are completing. These will often be Bash, Python, SSH, but can also be even cooler things like Docker, Kubernetes, AWS Batch, AWS ECS, Database Operations, file pushers, and more. Then there are also Sensors, which are nice and shiny ways of waiting for various operations, whether that is waiting on a file to appear, a record in a database to appear, or another task to complete.
Out of the box you get lots of niceness, including a nice web interface with a visual browser of your tasks, a scheduler, configurable parallelism, logging, watchers and any number of executors. As all of your configuration is written in code it is also extremely flexible. It can integrate with existing systems or stand on it's own.
There are any number of scientific workflow managers out there, and by the time I finish this article a few more will have popped into existence. Apache Airflow is my favorite, but you should shop around to see what clicks with you!
Computational Backends
I briefly touched on this earlier, but one of the perks that initially drew me to Apache Airflow is just how completely agnostic it is to your compute environment. You could have a laptop, a single server, a HPC cluster, or execute on the AWS or GCP. Airflow itself does not care. All you need to do is to map out your logic, make sure the data is available, and use whichever operator is appropriate.
Example CellProfiler Analysis Workflow
In this post I'm going to discuss the BBBC021 dataset and how I would organize and batch the analysis.
I decided to go for a simple setup, which is to use Apache Airflow with docker-compose and use the Docker operator to execute the CellProfiler analysis. Once you have your logic and workflow mapped out you could use any operator for any compute infrastructure, whether that is AWS ECS, or an HPC. My favorite lately has been Kubernetes, because it is not tied to any platform and can be used on AWS, GCP or in house. You can use Kubernetes to deploy your data visualization applications such as RShiny, Dash or Redash, and if you are using networked storage or S3 all your applications can access the same data!
Project setup
Let's setup our project directory structure!
mkdir CellProfiler-Apache-Airflow
cd CellProfiler-Apache-Airflow
mkdir -p data/dags
mkdir -p data/BBBC021/Week1
cd data/BBBC021/Week1
wget https://data.broadinstitute.org/bbbc/BBBC021/BBBC021_v1_images_Week1_22123.zip
find $(pwd) -name "*zip" | xargs -I {} unzip {}
# Clean up the zips, we don't need them anymore
find $(pwd) -name "*zip" | xargs -I {} rm -rf {}
cd ../
# Run $(pwd) to check where you are. You should be in /project/BBBC021
wget https://data.broadinstitute.org/bbbc/BBBC021/BBBC021_v1_image.csv
wget https://data.broadinstitute.org/bbbc/BBBC021/BBBC021_v1_compound.csv
wget https://data.broadinstitute.org/bbbc/BBBC021/BBBC021_v1_moa.csv
wget https://data.broadinstitute.org/bbbc/BBBC021/analysis.cppipe
wget https://data.broadinstitute.org/bbbc/BBBC021/illum.cppipe
# Let's create a data file ONLY for the week1 images, the first dataset
head -n 1 BBBC021_v1_image.csv > images_week1.csv
cat BBBC021_v1_image.csv | grep Week1_22123 >> images_week1.csvThis is mostly the Apache Airflow configuration from Bitnami. Bitnami is awesome and I use their configurations and images all the time. I made a few modifications to this one to bind our analysis dags, and also made a quick change so we could use the docker operator.
Dockerfile
We're going to use a custom Cellprofiler image to run our pipelines. Create a Dockerfile with this:
FROM cellprofiler/cellprofiler:3.1.9
RUN apt-get update -y; apt-get install -y unzip imagemagick
ENV TINI_VERSION v0.16.1
ADD https://github.com/krallin/tini/releases/download/${TINI_VERSION}/tini /usr/bin/tini
RUN chmod +x /usr/bin/tini
ENTRYPOINT [ "/usr/bin/tini", "--" ]
CMD [ "/bin/bash" ]Now we'll build out new CellProfiler image!
docker build -t cellprofiler .Grab the Docker Compose Configuration
Grab this file and save in your project root as docker-compose.yml.
version: '2'
services:
postgresql:
image: 'bitnami/postgresql:10'
- 'postgresql_data:/bitnami/postgresql'
environment:
- POSTGRESQL_DATABASE=bitnami_airflow
- POSTGRESQL_USERNAME=bn_airflow
- POSTGRESQL_PASSWORD=bitnami1
- ALLOW_EMPTY_PASSWORD=yes
redis:
image: bitnami/redis:5.0
volumes:
- 'redis_data:/bitnami'
environment:
- ALLOW_EMPTY_PASSWORD=yes
airflow-scheduler:
image: bitnami/airflow-scheduler:1
environment:
- AIRFLOW_LOAD_EXAMPLES=no
- AIRFLOW_DATABASE_NAME=bitnami_airflow
- AIRFLOW_DATABASE_USERNAME=bn_airflow
- AIRFLOW_DATABASE_PASSWORD=bitnami1
- AIRFLOW_EXECUTOR=CeleryExecutor
volumes:
- /var/run/docker.sock:/var/run/docker.sock
- ./data:/data
- ./dags:/opt/bitnami/airflow/dags
- airflow_scheduler_data:/bitnami
airflow-worker:
image: bitnami/airflow-worker:1
environment:
- AIRFLOW_LOAD_EXAMPLES=no
- AIRFLOW_DATABASE_NAME=bitnami_airflow
- AIRFLOW_DATABASE_USERNAME=bn_airflow
- AIRFLOW_DATABASE_PASSWORD=bitnami1
- AIRFLOW_EXECUTOR=CeleryExecutor
volumes:
- /var/run/docker.sock:/var/run/docker.sock
- ./data:/data
- ./dags:/opt/bitnami/airflow/dags
- airflow_worker_data:/bitnami
airflow:
image: bitnami/airflow:1
environment:
- AIRFLOW_LOAD_EXAMPLES=no
- AIRFLOW_DATABASE_NAME=bitnami_airflow
- AIRFLOW_DATABASE_USERNAME=bn_airflow
- AIRFLOW_DATABASE_PASSWORD=bitnami1
- AIRFLOW_EXECUTOR=CeleryExecutor
ports:
- '8080:8080'
volumes:
- /var/run/docker.sock:/var/run/docker.sock
- ./data:/data
- ./dags:/opt/bitnami/airflow/dags
- airflow_data:/bitnami
volumes:
airflow_scheduler_data:
driver: local
airflow_worker_data:
driver: local
airflow_data:
driver: local
postgresql_data:
driver: local
redis_data:
driver: localIf you're not used to containers this can get a little tricky, but one thing to note is that paths on the host are not necessarily the same as paths in the docker container, for example, our data directory could be anywhere on our host system, but is bound as /data on our container.
Put the docker-compose.yml file in your project directory and bring it up with docker-compose up. It may take some time to initialize. This is when I go make tea. ;-)
Once it's up you'll be able to access your Airflow instance at localhost:8080 with the default configuration.
AIRFLOW_USERNAME: Airflow application username. Default: user
AIRFLOW_PASSWORD: Airflow application password. Default: bitnamiThere won't be anything interesting here yet, because we don't have our analysis in place.
Grab the CellProfiler Analysis DAGs
First grab the illum dag. Place it in your dags folder. It can be named anything, what Airflow references is the dag_id, but I'll reference it as cellprofiler-illum-dag.py.
# dags/cellprofiler-illum-dag.py
from airflow import DAG
from datetime import datetime, timedelta
import string
import random
from airflow.utils import timezone
from airflow.operators.dagrun_operator import TriggerDagRunOperator
from airflow.operators.bash_operator import BashOperator
from airflow.operators.python_operator import PythonOperator
# Depending on which version of airflow you are on you will use either operators.docker_operator or providers.docker
from airflow.operators.docker_operator import DockerOperator
# from airflow.providers.docker.operators.docker import DockerOperator
from airflow.api.common.experimental.trigger_dag import trigger_dag
from airflow.sensors.external_task_sensor import ExternalTaskSensor
from airflow.api.common.experimental import check_and_get_dag, check_and_get_dagrun
import time
import os
from pprint import pprint
from airflow.utils.state import State
this_env = os.environ.copy()
this_dir = os.path.dirname(os.path.realpath(__file__))
default_args = {
'owner': 'airflow',
'depends_on_past': False,
'start_date': datetime(2019, 1, 1),
'email': ['[email protected]'],
'email_on_failure': False,
'email_on_retry': False,
'retries': 1,
'retry_delay': timedelta(minutes=5),
}
dag = DAG('cellprofiler_illumination', default_args=default_args, schedule_interval=None)
EXAMPLE_TRIGGER = """
{
"illum_pipeline" : "/data/BBBC021/illum.cppipe",
"analysis_pipeline" : "/data/BBBC021/analysis.cppipe",
"pipeline" : "/data/BBBC021/illum.cppipe",
"output": "/data/BBBC021/Week1/Week1_22123",
"input": "/data/BBBC021/Week1/Week1_22123",
"data_file": "/data/BBBC021/images_week1.csv"
}
"""
# Volumes are from the HOST MACHINE
illum = DockerOperator(
dag=dag,
task_id='illum',
retries=1,
volumes=[
# UPDATE THIS to your path!
'/path-to-project-on-HOST/data:/data'
],
working_dir='/data/BBBC021',
tty=True,
image='cellprofiler',
command=[
"bash", "-c",
"""cellprofiler --run --run-headless \
-p {{ dag_run.conf['illum_pipeline'] }} \
-o {{ dag_run.conf['output'] }} \
-i {{ dag_run.conf['input'] }} \
-data-file {{ dag_run.conf['data_file'] }} \
-c -r"""
]
)
def get_number_of_tasks(data_file):
"""
Parse the file to get the number of lines
The number of lines, minus 1 for the header
is the number of groups
:param data_file:
:return:
"""
file = open(data_file, "r")
number_of_lines = 0
for line in file:
number_of_lines += 1
file.close()
return number_of_lines - 1
def watch_task(triggers):
"""
This is only here for demonstration purposes
to show how you could dynamically watch the cellprofiler analysis DAG
:param triggers:
:return:
"""
print('-------------------------------------------')
print('Checking up on our dag...')
check_dag = check_and_get_dag(dag_id='cellprofiler_analysis')
dag_run = check_and_get_dagrun(check_dag, triggers[0].execution_date)
state = dag_run.get_state()
finished = State.finished()
unfinished = State.unfinished()
while state in unfinished:
time.sleep(10)
state = dag_run.get_state()
print('-------------------------------------------')
print('Dag run finished or dead')
pprint(dag_run.get_state())
def trigger_analysis(ds, **kwargs):
"""
Trigger the cellprofiler analysis DAG
We want one DAG run per row in the datafile, or -f / -l combo
:param ds:
:param kwargs:
:return:
"""
print('-------------------------------------------')
print("Here's the conf!")
pprint(kwargs['dag_run'].conf)
output = kwargs['dag_run'].conf['output']
data_file = kwargs['dag_run'].conf['data_file']
no_tasks = get_number_of_tasks(str(data_file))
triggers = []
print('-------------------------------------------')
print('Triggering our dag...')
for index, value in enumerate(range(1, no_tasks + 1)):
trigger = trigger_dag(
dag_id="cellprofiler_analysis",
replace_microseconds=False,
run_id="trig__{}__f_{}__l_{}".format(
timezone.utcnow().isoformat(),
value,
value
),
conf={
"pipeline": kwargs['dag_run'].conf['analysis_pipeline'],
"output": "{}/f-{}__l-{}".format(output, value, value),
"input": kwargs['dag_run'].conf['input'],
"data_file": data_file,
"first": value,
"last": value,
}
)
triggers.append(trigger)
trigger_analysis_task = PythonOperator(
dag=dag,
task_id='trigger_analysis',
provide_context=True,
python_callable=trigger_analysis
)
trigger_analysis_task.set_upstream(illum)And now our cellprofiler-analysis-dag.py.
# dags/cellprofiler-analysis-dag.py
from airflow import DAG
from datetime import datetime, timedelta
from airflow.operators.python_operator import PythonOperator
from airflow.operators.docker_operator import DockerOperator
import os
from pprint import pprint
this_env = os.environ.copy()
this_dir = os.path.dirname(os.path.realpath(__file__))
default_args = {
'owner': 'airflow',
'depends_on_past': False,
'start_date': datetime(2019, 1, 1),
'email': ['[email protected]'],
'email_on_failure': False,
'email_on_retry': False,
'retries': 1,
'retry_delay': timedelta(minutes=5),
}
dag = DAG('cellprofiler_analysis', default_args=default_args, schedule_interval=None)
analysis = DockerOperator(
dag=dag,
task_id='analysis',
retries=1,
volumes=[
# Volumes are from the HOST MACHINE
# UPDATE THIS to your path!
'/path-on-HOST/data:/data'
],
tty=True,
image='cellprofiler',
command=[
"bash", "-c",
"""cellprofiler --run --run-headless \
-p {{ dag_run.conf['pipeline'] }} \
-o {{ dag_run.conf['output'] }} \
-i {{ dag_run.conf['input'] }} \
--data-file {{ dag_run.conf['data_file'] }} \
-c -r -f {{ dag_run.conf['first'] }} -l {{ dag_run.conf['last'] }}"""
]
)
def gather_results(ds, **kwargs):
"""
Once we have the Cellprofiler results let's do something with them!
:param ds:
:param kwargs:
:return:
"""
pass
gather_results_task = PythonOperator(
dag=dag,
task_id='gather_results_task',
provide_context=True,
python_callable=gather_results
)
gather_results_task.set_upstream(analysis)Make sure that you update the DockerOperator volumes to match your local filesystem! Otherwise your analysis will not work!
Analysis Organizational Overview
What we have here are 2 separate DAGs, one for each CellProfiler Analysis.
The steps are:
- Process Illumination pipeline for ALL images
- Grab the datafile to see how many images we have
- Dynamically generate one CellProfiler submission per image.
- (Placeholder) Do something with our results!
You will notice that we are dynamically splitting the CellProfiler analysis. This will get our analysis done faster, which is increasingly important when you buy fancy robotic microscopes that generate oodles of data. Airflow takes care of the job queueing under the hood, so all we need to do is to figure out the logic of how we split the analysis. If you'd like to know more about how parallelism is handled in Airflow this is a great article.
Ok, that last one is just a place holder. I would imagine that you would want to do something with your results once you have them, such as put them in a database, fire off one or more analyses based on certain criteria, or do some post processing, but for now this is blank so you can wonder about the possibilities.
Passing arguments to our Analysis
This was not obvious to me when I started using Airflow, and now you get to hear all about it! ;-)
You pass arguments to a Airflow using the conf object or argument. Depending on the operator type this might look slightly different. If you are using the Python operator you access it as a dict, kwargs['dag_run'].conf, and if you are using Bash, or in this case Docker you access it as a templated variable {{ dag_run.conf['variable'] }}.
I will take you through how you use the Airflow web interface to trigger your DAG and pass in variables, but you can also trigger your DAGs using a REST API, through the Airflow CLI, or programatically using Python code. No matter how you trigger your DAG, you pass the configuration variables in as a JSON string.
A note on Docker Volumes
This is a little weird because we are using docker-compose to run Airflow, and then using the Docker Operator. Just keep in mind that when you use the Docker Operator you map your volumes using the paths on your HOST machine, not in the docker-compose containers. Your host machine is the machine you ran docker-compose up on.
For example:
analysis = DockerOperator(
...
volumes=[
# Volumes are from the HOST MACHINE
'/path-on-HOST/data:/data'
# NOT
# '/data:/data'
# Even though in our docker-compose instance the volume is bound as /data
],
...
)Analyze!
We're ready! Now that you have your dags (you need to have your dags) all setup we can start to analyze data!
Login at localhost:8080 with user user and password bitnami. You should see a screen that looks like this:
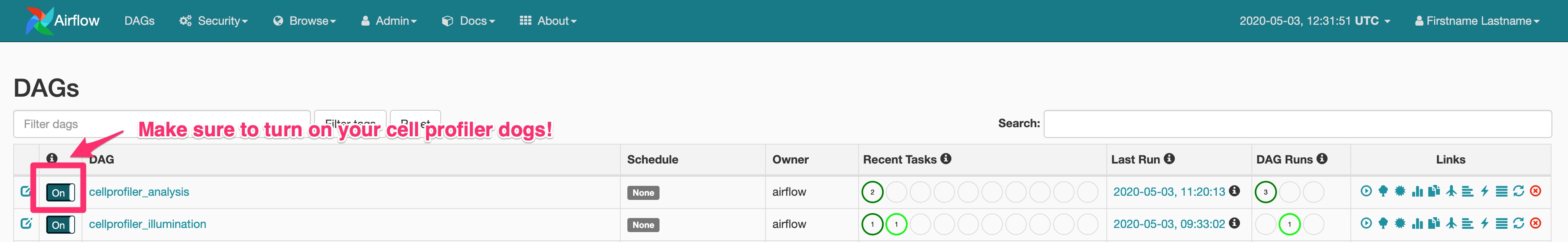
By default your dags will be off. Make sure to turn them on before proceeding to the next step!
Trigger your Analysis
Head to your main page at localhost:8080 and trigger the cellprofiler-illum DAG.
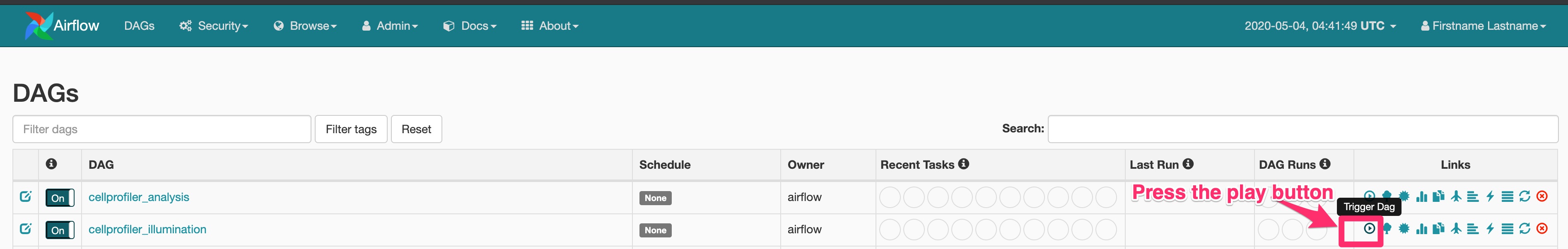
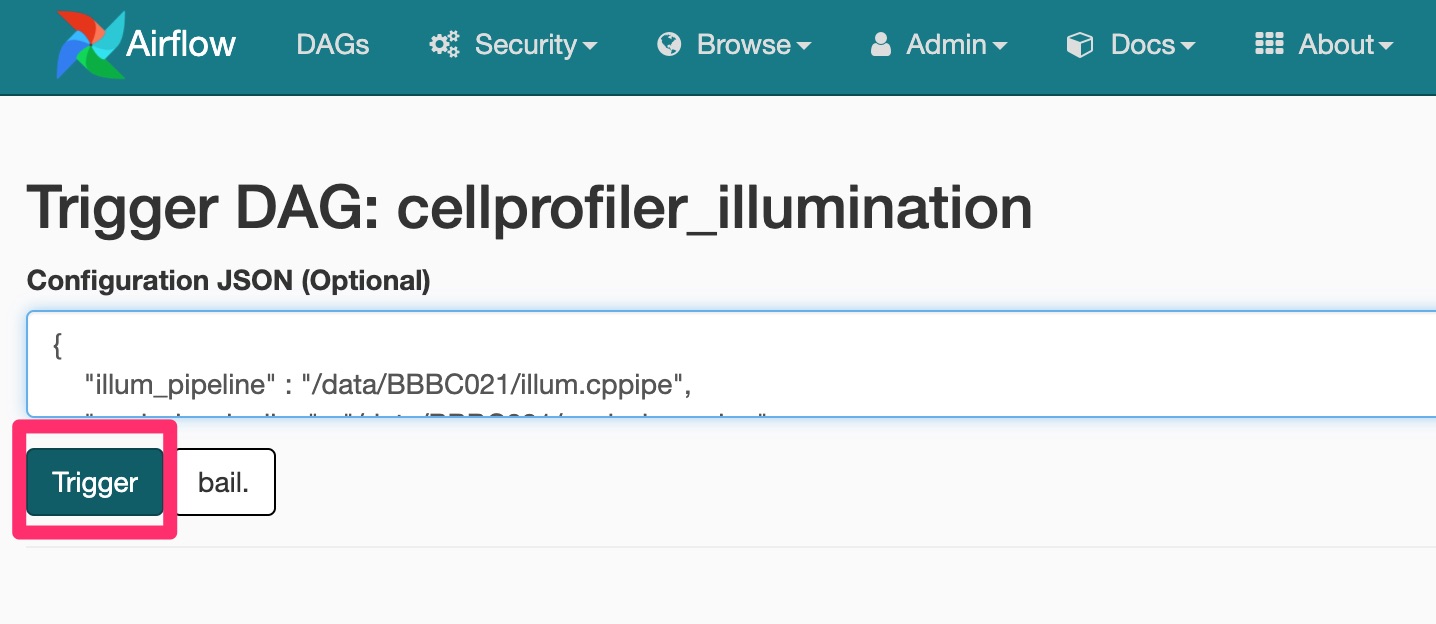
You will be prompted to add in some JSON configuration variables.
{
"illum_pipeline" : "/data/BBBC021/illum.cppipe",
"analysis_pipeline" : "/data/BBBC021/analysis.cppipe",
"pipeline" : "/data/BBBC021/illum.cppipe",
"output": "/data/BBBC021/Week1/Week1_22123",
"input": "/data/BBBC021/Week1/Week1_22123",
"data_file": "/data/BBBC021/images_week1.csv"
}
You'll be brought back to the main page and should see that your CellProfiler illumination analysis is running!

Here's a quick (~1 minute) video showing how you can navigate the Airflow interface to investigate your analysis.
Wrap Up
That's it! Let Airflow run and you will have your Illumination and Analysis pipelines running, all split nicely and something that isn't you babysitting keeping track of the job queue, logging, and success/failure rate. You can also integrate Airflow with any other system, such as a LIMS, reporting database, or secondary analysis workflow by using the REST API, the CLI, or code.
If you'd like to know more check out my website, or reach out directly at [email protected].
Acknowledgments
Special thanks to the Broad BioImage Repository for hosting the dataset used, along with Dr. Anne Carpenter, Beth Cimini, and Becki Ledford for extremely valuable feedback and editing!
Citations
https://data.broadinstitute.org/bbbc/BBBC021/
"We used image set BBBC021v1 [Caie et al., Molecular Cancer Therapeutics, 2010], available from the Broad Bioimage Benchmark Collection [Ljosa et al., Nature Methods, 2012]."
Work Together
Like what you see here? Check out my services page or email me directly at [email protected] to see how we can work together.

